Other Evolutionary Mechanisms
The theory of evolution itself has evolved significantly since 1858, when Charles Darwin and Alfred Russel Wallace published the first scientific papers identifying natural selection as the primary mechanism driving evolutionary change. Genetic recombination and mutation are now recognized as equally important mechanisms, since they provide most of the genetic diversity from which natural selection can “select” its favored varieties. Several additional mechanisms play similarly important roles in modern evolutionary theory. (The term mechanism, in this context, refers to a physical process that produces or explains some result.) Before examining whether evolution can explain the complexity and diversity of life, therefore, it will be worthwhile first to consider some of these other evolutionary mechanisms.
We can classify evolutionary mechanisms into two broad categories. Some evolutionary mechanisms produce new heritable traits in individual organisms, who may pass those new traits on to their descendants. Mutation and genetic recombination are two such mechanisms; additional examples from this category will be discussed below. Other evolutionary mechanisms operate on populations of a species to alter the frequencies (relative abundances) of traits within those populations.
Population-level Mechanisms
Let’s begin with the latter category: mechanisms that operate on populations. Natural selection is one such mechanism. In a population of finches living on the Galapagos islands, for example, natural selection may increase the frequency of beak shapes well-suited to the available foods and reduce the frequency of beaks that are disadvantageous, eventually eliminating the unfavorable beak genes from the population altogether. In addition to natural selection, several other mechanisms—including sexual selection, genetic drift, and gene flow—can also influence the frequencies of genes or alleles in a population. These additional mechanisms operate independently of natural selection and can even counteract the effects of natural selection in some circumstances. Here are brief explanations of these population-influencing mechanisms:
Sexual selection refers to the mating preferences of species that reproduce sexually. These preferences may be shaped by natural selection, for example when female birds prefer males with bright plumage indicative of good health. Nevertheless, sexual selection often counteracts the effects of natural selection, preserving and propagating disadvantageous genes that otherwise would be eliminated by natural selection. For instance, the extravagant tail feathers of the peacock encumber the bird, making it more vulnerable to predators; but peahens tend to mate more readily with ornately-adorned males, so the genes for beautiful feathers are preserved.
Darwin coined the term “sexual selection” in the original 1859 edition of his book On the Origin of Species, and he was the first scientist to recognize that sexual selection can contravene his own principle of natural selection. Citing a fellow naturalist who had noticed “how one pied peacock was eminently attractive to all his hen birds,” Darwin proposed sexual selection as the mechanism responsible for this apparent exception to the rule of natural selection.Charles Darwin, On the Origin of Species By Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life (London: Murray, 1859), 89. Available online hereGenetic drift occurs when the frequency of a gene in a population increases or decreases purely by chance, apart from the effects of natural selection. Genetic drift is most likely to occur in small, isolated populations. To see how this might happen, imagine a small flock of finches blown from the mainland to a remote island during a storm. Suppose, furthermore, that exactly ½ of the flock possesses the allele for some trait that makes little difference to their survival—a subtle color pattern on their feathers, say. (Whether the allele in question is dominant or recessive doesn’t matter for this example.) When the finches reproduce, their offspring might have a slightly different proportion of that allele: perhaps ⅔ of the baby birds will inherit it, or maybe only ⅓ will, just by chance.
After multiple generations, the allele for that specific color pattern might disappear from the population entirely, even though natural selection had nothing to do with its elimination. Or, just by chance, the allele may increase in frequency until every bird on the island has two copies of it—one from each parent—and all the alternative alleles have been eliminated. (When all of the alternatives are eliminated, an allele is said to be fixed in the population.) So long as no more birds from the mainland arrive at the island to reintroduce lost alleles into the isolated population, the castaways will continue to diverge genetically from their mainland counterparts. Eventually, the stranded finches may differ noticeably from the mainland species, even if the mainland environment is practically the same as the island environment and thus natural selection played no role in bringing about these genetic changes. Genetic drift can even contravene natural selection, eliminating advantageous traits or permanently fixing disadvantageous alleles in a population.
The fact that genetic drift can counteract natural selection poses serious risks for endangered species. When the total population of a species drops too low, genetic drift becomes inevitable, often resulting in the chance elimination of environmentally favorable alleles and the fixation of disadvantageous alleles. This leads to further decline in the population, resulting in faster genetic drift, and so on, until the species has little chance of survival. This vicious circle is known as the “extinction vortex.”Gene flow refers to the exchange of genes, via interbreeding, between separate populations of a species living in different environments or geographic locations. Returning to the insular flock of finches, for instance, gene flow will occur whenever a bird from the mainland finds its way to the island (or vice versa) and mates there, allowing genes from one population to “flow” into the other population. Gene flow tends to counteract the effects of genetic drift: whereas genetic drift causes the two populations to diverge, gene flow causes them to converge toward similar genetic traits.
In many circumstances, gene flow can also counteract the effects of natural selection. To illustrate, imagine that the primary food source on the island consists of nuts, so natural selection there favors larger beaks; but the primary food source on the mainland habitat consists of berries, which are readily consumed by small beaks. In that case, gene flow from the mainland to the island may periodically reintroduce the small-beak allele back into the island population so that it is never fully eliminated by natural selection.
All four of the population-influencing mechanisms mentioned above—natural selection, sexual selection, genetic drift, and gene flow—may help to explain trends in the heritable traits of diverse populations. However, none of those mechanisms can explain how new heritable features arise in the first place; they explain only changes in the frequencies of pre-existing traits. To explain the origin of novel traits, biologists turn to the other category of evolutionary mechanisms: processes that produce new heritable traits in individual organisms.
Genetic recombination and mutation are the most important—or, at least, the best-understood—mechanisms that produce new heritable traits. As discussed earlier in this chapter, recombination mixes and matches pre-existing alleles in ways that may yield novel traits within a limited range of variability. Mutation extends the range of variability by copying, deleting, or altering genes. There are limitations to what mutation can accomplish too, as we’ll see later in this chapter. However, not all heritable changes occur at the level of genes. Although mainstream evolutionary theory still regards genetic mutations as the primary source of novelty, biologists now think that other mechanisms must have played crucial roles as well. We’ll consider several proposed mechanisms in what follows.
Epigenetics and Evo-Devo
In addition to genetic recombination and mutation, numerous other mechanisms have been proposed to explain the emergence of new traits. Some of the most promising possibilities involve changes in the regulation of genes, rather than in the genes themselves. Soon after the genetic code was deciphered in the 1960s, geneticists discovered that DNA stores additional information besides the genes used to construct proteins. Most of a bacterium’s tiny genome consists of protein-coding genes; but, in more complex organisms, only a small fraction of the genome codes for proteins. For example, less than 2% of human DNA consists of protein-coding genes! Most of the remaining DNA was regarded, initially, as “junk DNA”—a useless vestige of our evolutionary history—but further discoveries have revealed (and are continuing to reveal) surprising functions for non-protein-coding segments of DNA.
In particular, much of the non-protein-coding DNA contains information used in gene-regulatory networks (GRNs), complex systems of chemical processes that control when and where genes are expressed. (A gene is expressed each time the cell uses its information to assemble a protein molecule.) An analogy may help to clarify the vital role of this regulatory DNA. Gene-regulatory networks are like the complex circuits in a computer processing system, while genes are like “user data” stored on a computer’s hard drive. The role of the non-protein-coding DNA involved in gene regulation is analogous to a computer’s operating system software, such as the Windows software on a PC or the Android software on a smartphone. The GRNs act like computer processors, using the operating system data stored in regulatory segments of DNA to control when and how the user data segments (protein-coding genes) are accessed.To extend the computer analogy a bit further, messenger RNA functions somewhat like a computer’s random-access memory, or RAM: it acts as a short-term-memory device, temporarily holding the data in a more accessible form. Biologist Eric Davidson, a pioneer in the analysis of GRNs, marveled at the “almost astounding” intricacy of GRNs found in the sea urchins he was studying. He described GRNs as “a network of logic interactions programmed into the DNA sequence that amounts essentially to a hardwired biological computational device.”Davidson, Genomic Regulatory Systems: Development and Evolution (New York: Academic Press, 2001), 54.
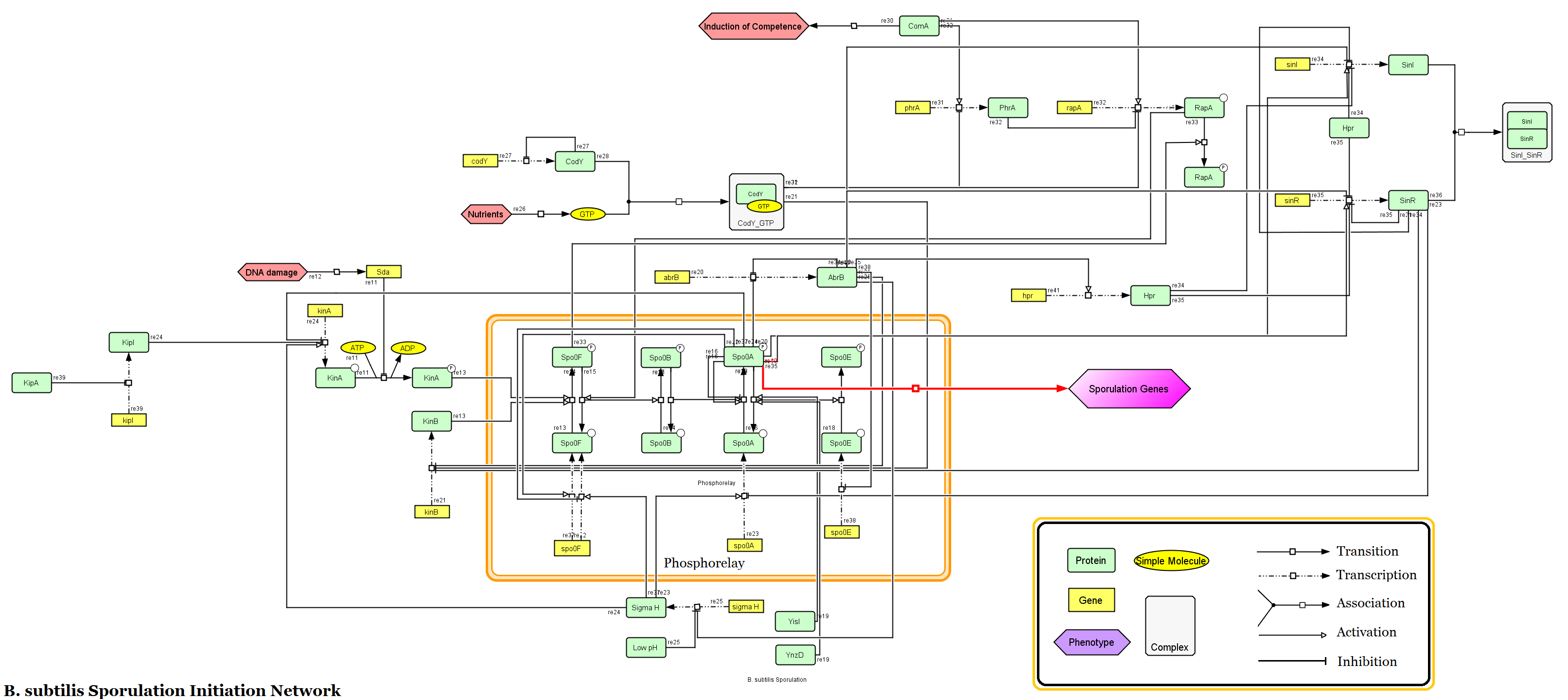
This diagram illustrates the operation of a relatively simple GRN controlling genes that initiate sporulation (the formation of a spore) in a common bacteria species named Bacillus subtilis. For an example of a stunningly complex GRN also found in bacteria, see this.
Though the mechanisms involved in GRNs are not yet fully understood, there are at least two ways in which they might be involved in the development of new heritable traits, as the relatively young scientific fields of epigenetics and evolutionary developmental biology have revealed:
To control the expression of genes, GRNs employ chemical “tags” or “marks” to label segments of DNA. Some of these chemical annotations act as switches to activate or deactivate a gene, switching the gene on or off as needed. Others tags regulate the frequency of gene expression, controlling how often the gene is used. Although these chemical tags are not part of the DNA itself, some of them are heritable: they can be passed from parents to offspring. Epigenetics is the study of this heritable information that is extraneous to DNA. Unlike the information encoded in DNA molecules, epigenetic information can be modified during an organism’s lifetime in response to environmental and behavioral factors. Thus, Lamarck was partly right after all: some behaviorally-induced changes can be inherited!
In a multicellular organism, developmental gene-regulatory networks (dGRNs) are involved in orchestrating growth and development processes as the organism matures to its adult form. Some dGRNs and their associated regulatory genes act as “master controls,” switching other genes on or off at specific times and locations in the body of a developing plant or animal. A mutation in one of these master genes can have drastic consequences. For example, experiments with fruit flies have shown that mutating a single master gene can cause the flies to develop without eyes. More surprisingly, researchers found that activating this so-called eyeless gene at the wrong times and places during a fruit fly’s development caused the fly to grow eyes in unusual positions, including on its legs, wings, and antennae!Georg Halder, Patrick Callaerts, and Walter J. Gehring, “Induction of Ectopic Eyes by Targeted Expression of the eyeless Gene in Drosophila,” Science 267, no. 5205 (March 1995): 1788-92. <https://www.science.org/doi/10.1126/science.7892602> (These extra eyes were not functional, however, since they lacked the nerve circuitry that would connect them to the fly’s brain.)
The emerging field of evolutionary developmental biology, nicknamed evo-devo, has begun to explore the possibility that major transitions in the evolution of life—especially, the emergence of radically new body structures (new “body plans” or Bauplans)—may have involved reprogramming of developmental GRNs. It is not yet clear how such reprogramming could occur, since mere mutations in the DNA would not be sufficient to restructure a whole GRN, much less construct a new GRN from scratch. Nonetheless, many biologists are optimistic that evo-devo research eventually will reveal developmental mechanisms that could plausibly explain major evolutionary transitions. Not all biologists share this optimism. For a critical discussion of evo-devo, see chapter 5 of Michael Denton’s book Evolution: Still a Theory in Crisis (Seattle: Discovery Institute Press, 2016), 83-101.
Epigenetics and evo-devo research have also suggested evolutionary mechanisms that don’t directly involve genes at all. As it turns out, DNA is not the only vehicle by which heritable traits are transmitted from parents to offspring. Many other cellular structures and molecules also carry hereditary information, especially structural information important to embryonic development. For example, structural patterns in cell membranes are copied directly from parent to offspring cells during cell division, independently of DNA. These heritable features of the cell membrane include patterns of ion channels, which transport charged ions through the membrane, producing electric fields that help to regulate developmental processes. Another heritable feature of the cell membrane, the so-called sugar code, consists of highly complex arrangements of sugar molecules attached to the membrane surface. The pattern of sugar molecules acts as a sort of digital code analogous to the genetic code stored in DNA. In multicellular organisms such as plants and animals, the sugar code and the electric fields created by ion channels both provide positional information to guide the formation of macroscopic structures during embryonic development.
Additionally, in animal cells, a structure called the centrosome is duplicated directly during cell division. It too carries heritable information distinct from DNA. The centrosome is primarily responsible for organizing complex arrays of protein filaments known as microtubules, which comprise part of the cytoskeleton (the cell’s internal “skeleton” or framework). Like the cell membrane, the centrosome and cytoskeleton help to guide embryonic development, while also serving many important purposes within the individual cell. So do maternal RNA molecules stored in the pre-fertilized egg cell, along with numerous other factors extraneous to the organism’s DNA.
All of these heritable features—the sugar code, patterns of ion channels, structures in the centrosome, etc.—act in concert with genetic information to guide the embryonic development of animals, influencing the shapes and positions of limbs, organs, and other anatomical structures. Thus, changes in any of those diverse forms of non-DNA-based information might give rise to new traits, perhaps even contributing to large-scale evolutionary transitions. Indeed, macroevolutionary transitions must have involved not only genetic mutations but significant alterations in non-genetic forms of heritable information as well, assuming such transitions occurred at all.For further discussion of these heritable features and their significance for evolutionary theory, see Stephen C. Meyer, Darwin’s Doubt: The Explosive Origin of Animal Life and the Case for Intelligent Design (New York: HarperOne, 2013), Chapter 14. These possible mechanisms of evolution are not well understood, but some biologists think that new variants in the sugar code, membrane ion channels, and centrosome structures (etc.) might be introduced via copying errors or other processes analogous to DNA mutation.On the other hand, there are good reasons to doubt that such processes could explain major evolutionary innovations, as discussed later in this chapter. See also Stephen C. Meyer, Darwin’s Doubt: The Explosive Origin of Animal Life and the Case for Intelligent Design (New York: HarperOne, 2013), Chapter 14 (especially the section on “Epigenetic Mutations”) and Chapter 16.
Symbiogenesis
A number of other possible evolutionary mechanisms have also been proposed. Most are highly speculative and controversial. However, one further idea is worth mentioning, as it has gained widespread acceptance as an explanation for some of the complex structures within cells. All living cells contain a variety of specialized structures called organelles, which are in some ways analogous to bodily organs: each organelle performs a specific function within the cell, just as organs (heart, lungs, etc.) perform specific functions in an animal’s body. The cytoskeleton and cell membrane, mentioned above, are important organelles found in all living cells.Although all living cells have a cytoskeleton, not all cytoskeletons include a centrosome. That feature is unique to animal cells, providing an additional source of heritable structure not found in other kinds of organisms. In many cells, the largest organelle is the nucleus, which contains most of the cell’s DNA. (Some DNA is also found in other organelles, as we’ll see in a moment.) However, bacteria and archaea do not have a nucleus. These prokaryotes, as they are called, have other organelles; but their DNA is not enclosed within any structure apart from the cell membrane that surrounds the whole organism. Eukaryotes, in contrast, are defined as organisms whose cells do contain a nucleus. All known organisms except bacteria and archaea belong to this category.Modern taxonomic conventions classify eukaryotes, bacteria, and archaea as the three domains of life. Domains are the highest taxonomic category for living organisms, above kingdoms, phyla, etc.
According to a popular (albeit speculative) hypothesis known as the theory of symbiogenesis, some of the organelles found in eukaryotic cells originated as independent prokaryotic organisms. For example, chloroplasts—the green-colored organelles that carry out photosynthesis in the cells of plants and algae—may have originated as independently-living prokaryotic organisms. Once upon a time in the remote past, the theory suggests, a green bacteria-like organism capable of photosynthesis was engulfed by a larger cell. Rather than promptly digesting the little green cell, however, the larger cell instead digested sugars produced by photosynthesis in the little cell. Thus, the two cells entered into a mutually beneficial, symbiotic relationship. The larger cell received food from the smaller cell, while the little green cell (which could make its own food via photosynthesis) found shelter and protection inside the larger cell. Both cells multiplied, and their descendants eventually evolved into the various plant and algae species we see today.
Similar stories have been told to explain the origin of mitochondria—organelles that use chemical energy from sugars, fats, and other fuel sources to charge up a special molecule called adenosine triphosphate (ATP), which acts as a rechargeable battery to power many other processes throughout the cell. Interestingly, both mitochondria and chloroplasts contain their own DNA, distinct from the DNA in the cell nucleus. This lends plausibility to the symbiogenesis explanation, at least for these two organelles, though it is more difficult to envisage similar explanations for the many other organelles in complex eukaryotic cells.
The modern theory of evolution incorporates all of the mechanisms discussed above to construct an elaborate account of the history of life. According to this theory, biological life began with a relatively simple, single-celled organism. (Or, perhaps, it began with an even simpler self-replicating system that wouldn’t be considered a “cell.” We’ll return to that suggestion later in this chapter.) For billions of years, mutations gradually diversified the traits of that organism’s descendants. Symbiogenesis, GRN modification, and perhaps other processes may have contributed to the diversification as well. Eventually, some of the organism’s descendants developed traits that allowed them to reproduce sexually; thereafter, genetic recombination also played a role in producing new variations. All the while, genetic drift carried isolated populations toward even greater diversity, and natural selection eliminated traits that were not well-suited to each environment, thereby steering evolution toward the specific varieties of life we see today. Did all that really happen? We’ll consider evidence for and against the theory in the remaining pages of this chapter.